Reinke’s Edema (RE) is a laryngeal lesion related to excessive tobacco smoking, voice overuse, and laryngopharyngeal reflux. Although the risk of malignancy has been considered low in literature, RE is classified among precancerous lesions.
ObjectivesWe investigated DNA Copy Number Alterations (CNAs) in specimens of RE and its potential association with malignant progression.
MethodsWe used array-based comparative genomic hybridization (aCGH, Agilent 4 × 180 K platform) to study eight RE cases. All patients were heavy tobacco users for at least 30 years, and none of them progressed to cancer in the follow-up (>8 years). Two RE presented mild dysplasia, one moderate dysplasia, and no histological alterations were found in the remaining five cases. CNAs were compared with the Database of Genomic Variants (DGV) and genes mapped on altered regions had their functions annotated.
ResultsSix of eight patients showed different rare copy number alterations on chromosomes 2q37.3, 4q13.1, 4q13.3, 7q11.22, 10p14, and 13q34. A gain of the whole chromosome 8 were detected in one case. Of interest, four of eight RE cases showed copy number imbalances involving genes previously described in several tumor types (RASA3, COL6A3, LINC00707, LINP1, SMR3A, and SMR3B).
ConclusionThe genomic imbalances herein found in RE have the potential to contribute to the phenotype but with limited or no risk of cancer. A long-term follow-up in a large series of patients could clarify the mechanisms involved in the malignant progression of RE.
Level of evidence4.
Reinke’s Edema (RE) is a chronic laryngeal lesion related to excessive tobacco smoking showing high prevalence in women. The voice gradually becomes hoarse and with a lower pitch.1,2 Although controversial, Reinke’s edema has been classified as a preneoplastic lesion.3–6 In 54 smokers with RE, Martins et al. reported that RE could harbor dysplasia and carcinomas.6 Also, Avila et al.7 described an association between premalignant lesions and RE. These findings supported the hypothesis that the ER may be a preneoplastic lesion.
The molecular characterization of these lesions is poorly explored. Large-scale expression analysis in 11 RE compared with 17 laryngeal polyps revealed 65 differentially expressed genes (40 under-expressed and 25 over-expressed).8 Interestingly, 19 of these genes were associated with gastroesophageal reflux disease. Proteomic analysis of human vocal fold fibroblasts, cultured in a medium with cigarette smoke extract, identified an increased production of proteins related to oxidative stress, as well as hyaluronan.9
Array-based Comparative Genomic Hybridization (aCGH) is a molecular cytogenetic procedure used to evaluate the global DNA copy number changes across of whole genome of a tumor (test sample) relative to the normal tissue.10 Copy Number Alterations (CNAs) are structural alterations that include gains, amplifications, losses, and deletions generated by breakpoints, genomic rearrangements, and reassembly of DNA sequences. CNAs may include a single or few oncogenes and tumor suppressor genes with potential impact on cancer development and tumor progression.11–13 Since RE could be a potential risk factor for tumor initiation, we used the array-based Comparative Genomic Hybridization (aCGH) methodology to evaluate patterns associated with subsequent malignancy. We are not aware of other studies assessing global copy number alterations in Reinke’s edema.
MethodsPatients and samplesEight adult patients (mean age at diagnosis was 46.5 years) diagnosed with RE (2011–2014), confirmed by video laryngoscopy, were subjected to laryngeal microsurgery. The diagnosis was confirmed by histopathological evaluation. Exclusion criteria were patients with laryngeal diseases other than RE, or with the presence of infectious or chronic diseases affecting the larynx or airways, or previous diagnosis of laryngeal cancer or in situ carcinoma. Epithelial alterations were classified using the grading systems of dysplasia in the head and neck, according to Fleskens and Slootweg.14 Medical records were consulted to retrieve the smoking habits, demographic information, and clinical follow-up data.
The study was approved by the Research Ethics Committee at the Faculty of Medicine, UNESP, Sao Paulo, Brazil (REB #4345-2012). All patients signed informed consent before sample collection.
Array-based comparative genomic hybridization (aCGH) profilingDNA was isolated using the standard phenol-chloroform method, followed by quantification (Nanodrop™ 8000, Thermo Fisher Scientific).15,16 Copy number data was generated using the 4 × 180 K aCGH platform (Agilent Technologies, Santa Clara, California, USA). DNA digestion, labeling, and hybridization were performed according to the manufacturer’s instructions (Agilent Technologies). A commercial human genomic DNA (Human Reference DNA, Male and Female, Agilent Technologies) was used as normal reference.
Data were extracted with Feature Extraction software v10.1.1.1 (Agilent Technologies), and the analyses were performed using CytoGenomics software v5.1 (Agilent Technologies) with the following parameters: detection algorithm ADM-2 (Aberration Detection Method 2), threshold 6.0; Fuzzy Zero, GC correction, diploid peak centralization, and at least four consecutive probes showing CNAs with log2 ratio ≥0.25 or ≤−0.25 for gains or losses, respectively. The alterations were visually inspected, with poor quality data excluded. CNAs were compared with the Database of Genomic Variants (DGV), and a database of 100 healthy individuals of the Brazilian population.17,18 CNAs with the same size and type identified in <0.05% of DGV patients and <5% of the Brazilian database were classified as rare. In addition, genes mapped in regions of copy number gains or losses had their functions annotated using the National Center for Biotechnology Information (NCBI).19 Microarray data have been deposited in NCBI’s Gene Expression Omnibus (GEO) and are accessible through GEO Series accession number GSE176430.20
ResultsOur sample set was composed of four men and four women. Histopathological analysis showed mild dysplasia in two RE (patients 3 and 5), moderate dysplasia in one RE (patient 6), and no histological alterations in the remaining five cases. All patients were heavy tobacco users (25–135 packs per year) for at least 30 years (except one patient aged 31 years). Six patients reported moderate or low alcohol consumption and two did not drink alcohol (cases 4 and 5). Gastroesophageal reflux disease was found in two patients (cases 2 and 8). Three patients presented personal and/or family history of cancer. Patient 3 presented a personal history of malignant bladder neoplasia and relatives with breast (mother) and bladder cancer (cousin). Family history of prostate cancer (father) and gynecological malignant neoplasia (sister) was reported by patient 7. Patient 8 reported an uncle and cousin with cancer of an unspecified site. The patients had at least eight years of follow-up after the surgical procedure, and no malignant disease was found. Table 1 summarizes the clinical data of our dataset.
Clinical characterization and rare copy number alterations in Reinke’s edema patients.
| Case | Age/gender | Type of alteration | Chromosome | Starte | Ende | Number of probes | Genes |
|---|---|---|---|---|---|---|---|
| 1 | 31/M | Loss | 4q13.1 | 60403039 | 60482470 | 5 | ‒ |
| 2a | 47/F | Gain | 13q34 | 114747920 | 114773099 | 4 | RASA3 |
| 3b,d | 58/M | Loss | 10p14 | 6677672 | 6845446 | 9 | LINC02648, LINP1, LINC00706, LINC00707, |
| 4 | 49/F | Gain | 2q37.3 | 237826846 | 238245191 | 22 | COPS8, COL6A3 LOC93463 |
| 5b | 47/F | Loss | 7q11.22 | 69520507 | 69563017 | 5 | AUTS2 |
| 6b,d | 51/M | ‒ | ‒ | ‒ | ‒ | ‒ | ‒ |
| 7c | 46/F | Loss | 4q13.3 | 71162798 | 71283216 | 10 | CABS1, SMR3A, SMR3B, OPRPN |
| Gain | 8p11.21-p23.3 | 1522999 | 42738998 | 2418 | Several genes | ||
| Gain | 8q11.1-q24.3 | 46943457 | 146066584 | 5312 | Several genes | ||
| 8a | 43/M | ‒ | ‒ | ‒ | ‒ | ‒ | ‒ |
M, male; F, female.
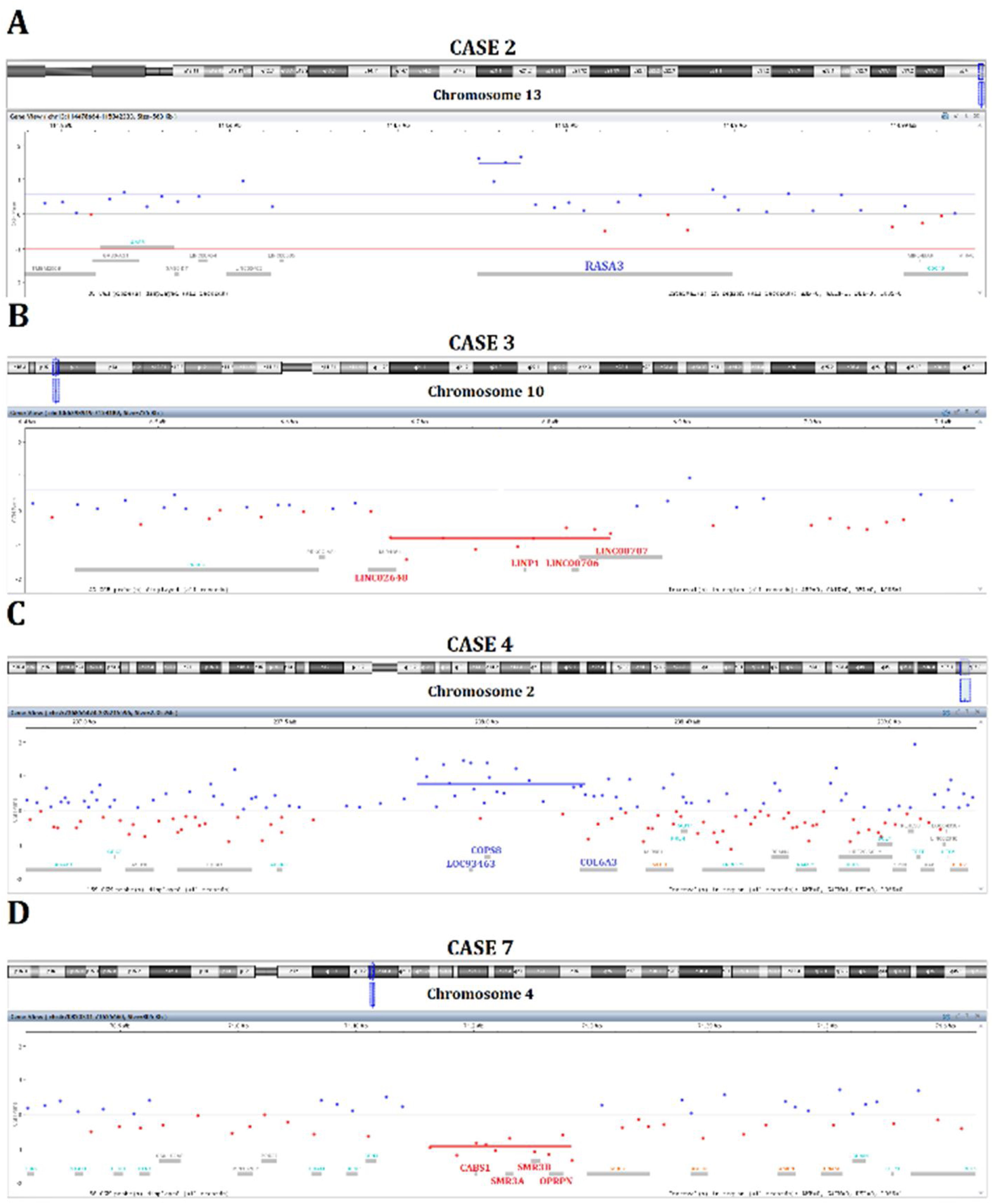
Rare copy number alterations were detected in six cases involving different chromosomal regions (Table 1). One patient (case 2) exhibited a gain of 13q14 (Fig. 1A). Case 3 presented a loss of 10p14 affecting a cluster of long intergenic non-coding RNAs (Fig. 1B). A gain of 2q37.3 was identified in case 4 encompassing three genes (Fig. 1C). Interestingly, case 7 showed gains of the whole chromosome 8 and loss of 4q13.3 (Fig. 1D). Case 1 also had a deletion of 4q13, but no genes mapped to it. Two cases (patients 6 and 8) presented only common genomic alterations.
DiscussionTo our knowledge, this is the first study describing genomic alterations in RE. The genomic revealed low genomic instability and a small number of rare copy number alterations. Two cases have no significant rare CNAs, and one (case 1) presented a loss of 4q13.1 where no known gene is mapped. However, in particular cases, we found chromosomal imbalances affecting genes with potential significance to the disease.
Gains of 13q14, where RASA3 gene is mapped, were detected in case 2. The protein encoded by this gene is a member of the GAP1 family that acts as a RAS-GTPase-Activating Protein (GAPs) and negatively regulates the RAS pathway. The functional role of RASA3 in proliferative disorders remains to be established. However, RASA3 can inhibit RAP1, resulting in invasion and metastasis in ovarian and colorectal cancer.21–23 RASA3 was suggested as a tumor suppressor in comparative studies of dogs and human cancers.24
We found a loss of 10p14 affecting a cluster of four long intergenic non-coding RNAs; LINC02648, LINC00706, LINC00707, and LINP1 (case 3). Long non-coding RNAs (lncRNAs) have been involved in gene regulation, such as gene transcription, genomic imprinting, mRNA splicing, and protein activity interfering with signaling pathways.25 Recent studies have suggested that the long intergenic non-protein coding RNA 70 (LINC00707) acts as an oncogene in colorectal, breast, hepatocellular, renal, and lung carcinomas.26–31 Additionally, LINP1 overexpression has been implicated in the progression of several cancers, such as acute myeloid leukemia, prostate, and breast neoplasms.32–34
Case 4 presented gains of COPS8, COL6A3, and LOC93463 mapped in 2q37.3. COPS8 encodes one subunit of COP9 signalosome, a highly conserved protein complex, which plays a regulatory role on multiple signaling pathways. In gastric tumor cells, COPS8 is targeted by the miR-146a.35 COL6A3 gene encodes the alpha 3 chain of type VI collagen, and it is an essential component of the extracellular matrix. Previous studies reported that in stroma cells from colorectal carcinomas, COL6A3 was upregulated and associated with a worse prognosis.36 Interestingly, Hsa-cir-0006401 is a circular RNA (circRNA) derived from its host gene COL6A3 (exons 2–4). The non-coding RNAs named circRNAs are generated by a back-splicing mechanism with a role in various cellular processes and tumorigenesis.37–39 Zhang et al.39 reported an increased expression level of Hsa-cir-0006401, which encodes the 198-aminoacids peptide, and is a potential marker of metastatic colorectal cancer. Gains of 2q37 were also described in laryngeal carcinomas.40 Interestingly, the surface area of one transglottic tumor from this cohort,40 presented gains of COPS8 and COL6A3 (mapped on 2q37), the same genes altered in case 4.
Loss of 7q11.22 where is mapped AUST2 (autism susceptibility candidate 2) gene was detected in one RE with moderate dysplasia (patient 5). Disruption of AUT2 by different mechanisms, including copy number alterations, has been associated with developmental delay and intellectual disability.41 The phenotype of the patients with this syndrome is variable from severely to moderately affected. Although patient 5 had no apparent clinical features of this syndrome, further neurological evaluation and molecular assays in normal tissues should clarify the significance of AUST2 loss in our patient.
The only case of our series that presented large genomic imbalances was patient 7 (RE with moderate dysplasia), showing gains covering the whole chromosome 8 and loss of 4q13.3 (CABS1, SMR3A, SMR3B, OPRPN genes). Among the genes mapped on 4q13.3, CABS1 is a spermatic-specific protein, and OPRPN is a proline rich protein family, both with unknown function in proliferative disorders. The Submaxillary gland Androgen-Regulated protein 3A (SMR3A) and SMR3B belong to the family of opiorphins. Increased expression of SMR3A was reported in oropharyngeal squamous cell carcinoma, while decreased expression was found in adenoid cystic carcinoma.42,43 The trisomy 8 found in malignancies, such as acute myeloid leukemia and myelodysplastic syndrome, may derive from a constitutional trisomy 8 mosaicism.44 Trisomy 8 found as constitutional mosaicism, characterizes the Warkany syndrome. This rare condition presents variable phenotypes ranging from mild dysmorphic features to severe malformations.45 Our patient had no clinical features of Warkany syndrome and, after a long-term follow-up (8 years), had no history of leukemia or myelodysplastic syndrome.
Overall, six RE presented significant rare copy number alterations, and four cases showed genomic imbalances with a potential role in the disease. Although the genomic profile found in our cases presented a heterogeneous pattern, all patients were heavy smokers, and therefore, these alterations probably are not directly related to the smoking habit. Also, after a long-term follow-up (at least 8 years), none of them had cancer, supporting the rarity of disease progression reported in the literature.
ConclusionOur study demonstrates that the genomic alterations found in Edema’s Reinke cases may have no association with malignant progression.
Conflicts of interestThe authors declare no conflicts of interest.
The authors would like to thank the patients who kindly agreed to participate in this study. We also thank the Botucatu Clinical Hospital Faculty of Medicine, São Paulo State University (UNESP), Botucatu, SP, Brazil and International Research Center, A.C. Camargo Cancer Center, Sao Paulo, SP, Brazil where samples were collected and analyzed, respectively.
Peer Review under the responsibility of Associação Brasileira de Otorrinolaringologia e Cirurgia Cérvico-Facial.